Intro
Introduction
Description
Description
Reasoning
Reasoning
Approach
Approach
Imports
Imports
Code
Here we can see that we will be storing a lot of variables, mostly from the fit function so that we can store models and more vital variables so that they can be used in the prediction method later on in the class. Each stage is stored as to be able to give a comprehensive look into each of the components, following a fit. This can be useful when trying to see each of the different steps and models used and feature importances for each model.
Storing all these features gives a great opportunity to further develop the class to include an internal score function, model feature importances and other vital pieces of information acquired throughout the fitting phase.
Storing all these features gives a great opportunity to further develop the class to include an internal score function, model feature importances and other vital pieces of information acquired throughout the fitting phase.
Model Validations
When developing an API, in order to ensure that the correct parameters are being entered, assertions are great for ensuring this is the case. I added a model validation step to ensure that the models could in fact be used within the stacking phase. Here I have asserted that the models do in fact have a "predict_proba" or "predict" function. I then append a list which is used to determine which function to use for each model dependant on their index, as well as pass a note to the user if their is no "predict_proba" (a preferred prediction) and there is a "predict" function, that the latter will be used. I look to add mode validations to the class to ensure that the class is as robust as possible, after users have used the class, I will be able to make more informed adjustments to the assertions and directions of use.
Static Predictor method
Here we have a static method that runs the prediction of any of the given model. I have this method as there are a number of predictions taking place and of different variants requiring a conditional filter, and due to this, reducing this to a function allows for a much smaller class.
Fitting the Stacker
The fit method within the stacker is understandably the biggest method of them all seeing as this is where the stacking and fitting of all the models take place. During this method, we can see that we save a lot of the variables that were initialised at the beginning.
Here we start by re-initialising the class. This will remove any variables or models from any previous fits of the model as to ensure a fresh fit is modelled. I then ensure that the X and y variables passed into the fit function are pandas dataframes, as these are easier to handle and have useful functions as mentioned earlier. If they aren't in the desired format, then these are converted to pandas dataframes.
Next, the method looks at the model feature indices. This variable indicates, for each model, which features should be selected from X, given the indices passed. These are easily filtered with an ".iloc[]" method within pandas. If this parameter is not given, then the model will assume that all features should be selected and does so.
Next, the model begins the meta-feature generation phase and enters a loop across all the models, selected features and the prediction method in order to generate the predictions and appends a dataframe where all the meta-features are stored. Within this loop is a conditional argument to take into account whether blending is to take place or not as the blending process is different as this loops through a KFold of the training data to generate predictions.
Once all the meta-features are generated, they are appended to the main training data and the models that were trained to generate the features are stored. The stacker model is then fit with the new training and meta-feature data and stored within the object for predictions later.
Next, the method looks at the model feature indices. This variable indicates, for each model, which features should be selected from X, given the indices passed. These are easily filtered with an ".iloc[]" method within pandas. If this parameter is not given, then the model will assume that all features should be selected and does so.
Next, the model begins the meta-feature generation phase and enters a loop across all the models, selected features and the prediction method in order to generate the predictions and appends a dataframe where all the meta-features are stored. Within this loop is a conditional argument to take into account whether blending is to take place or not as the blending process is different as this loops through a KFold of the training data to generate predictions.
Once all the meta-features are generated, they are appended to the main training data and the models that were trained to generate the features are stored. The stacker model is then fit with the new training and meta-feature data and stored within the object for predictions later.
Predictions
Here we have the prediction method. This is partitioned into 2 parts, one where blending has been applied and one where blending isn't. This method takes the trained models that were built during the "fit" method and builds the metafeatures. If blending has been applied, we will have as many models as there are folds in the data. The default value for the folds is 5, therefore 5 models will then be used to generate the metafeature. Their results will then be averaged out to provide the metafeature for that model type. This will be repeated for each of the models that are to be stacked.
Once all the metafeatures have been generated, the pre-trained stacker will then take the new X data and the metafeatures generated and produce predictions which are then returned.
Once all the metafeatures have been generated, the pre-trained stacker will then take the new X data and the metafeatures generated and produce predictions which are then returned.
Test Script and Examples
Here we have a test script to assess the results of the model. The data used to test the stacker is from sklearn's datasets module. The data is the 'Breast cancer wisconsin (diagnostic) dataset'. This data is a classification datasets in that the targets are binary, either breast cancer is malignant (1) or if it is benign(0). There are 212 malignant cases and 357 benign cases. This dataset consists of 569 cases and 30 numerical features to describe each case. features include things like:
The model used to evaluate the performance are:
The data can be found here (https://goo.gl/U2Uwz2).
- 'mean radius'
- 'mean texture'
- 'mean perimeter'
- 'mean concave points'
- 'mean symmetry'
- 'mean fractal dimension'
The model used to evaluate the performance are:
- Random Forest
- Logistic Regression
- Decision Tree
- Ridge Regression
- Stacker Model
The data can be found here (https://goo.gl/U2Uwz2).
Results
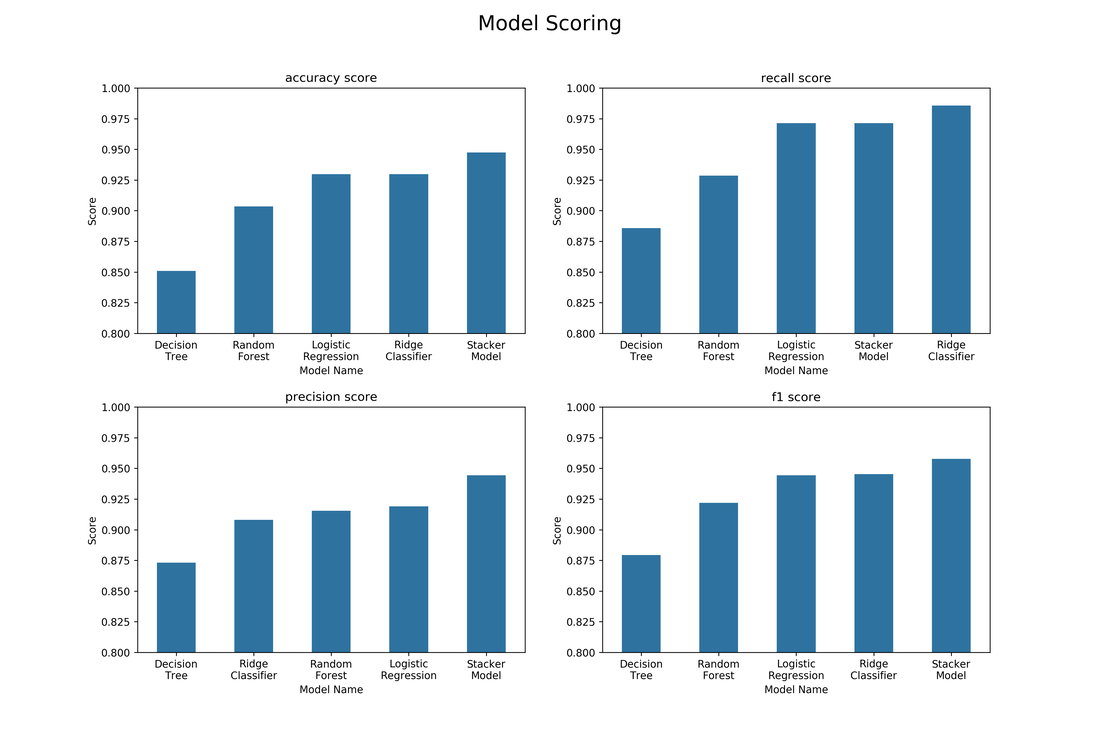
Having run the script above we can have a look at the results. He we can see that the stacker model performs better than all the other models when evaluating the accuracy, precision and f1 score of the model. The ridge classifier, however, performs the best in terms of recall but has a very poor precision score. Based on these scores, we can see that the stacker performs better in general. The model is able to take advantage of the other models by ensembling their scores. The f1 score is an evaluation metric that gives a good all round performance of a model and here we can see that the stacker model performs the best. The blending as well as the ensembling provides the strength in this approach as blending helps generalise the model and ensembling helps improve the predictive power of the model.
Final Thoughts, Ideas and Next Steps...
Overall, we can see that stacking does in fact provide better results but this is then accompanied by increased complexity. As mentioned earlier, model stacking is great for competitions where every small percent is valuable in differentiating yourself from the competition. Hopefully this will provide an easy approach to stacking so that many people can start using it with models from scikit-learn. It can be noted that this is in-fact built with scikit-learn models in mind, however, this could be used with other models granted that they have the same methods that are required. With that in mind, you could stack a bunch of stackers together as this implementation has the relevant methods to do so.
Moving forward, I would like to add more descriptive and diagnostic methods that provide more information about the models used inside the stacker model. Methods that I would like to include are scoring methods, feature importance methods and coefficient methods etc. I would also like to add more assertions and testing in order to ensure the stacker model is as robust as possible.
Moving forward, I would like to add more descriptive and diagnostic methods that provide more information about the models used inside the stacker model. Methods that I would like to include are scoring methods, feature importance methods and coefficient methods etc. I would also like to add more assertions and testing in order to ensure the stacker model is as robust as possible.
To install the package onto your local machine you can find it within pip.